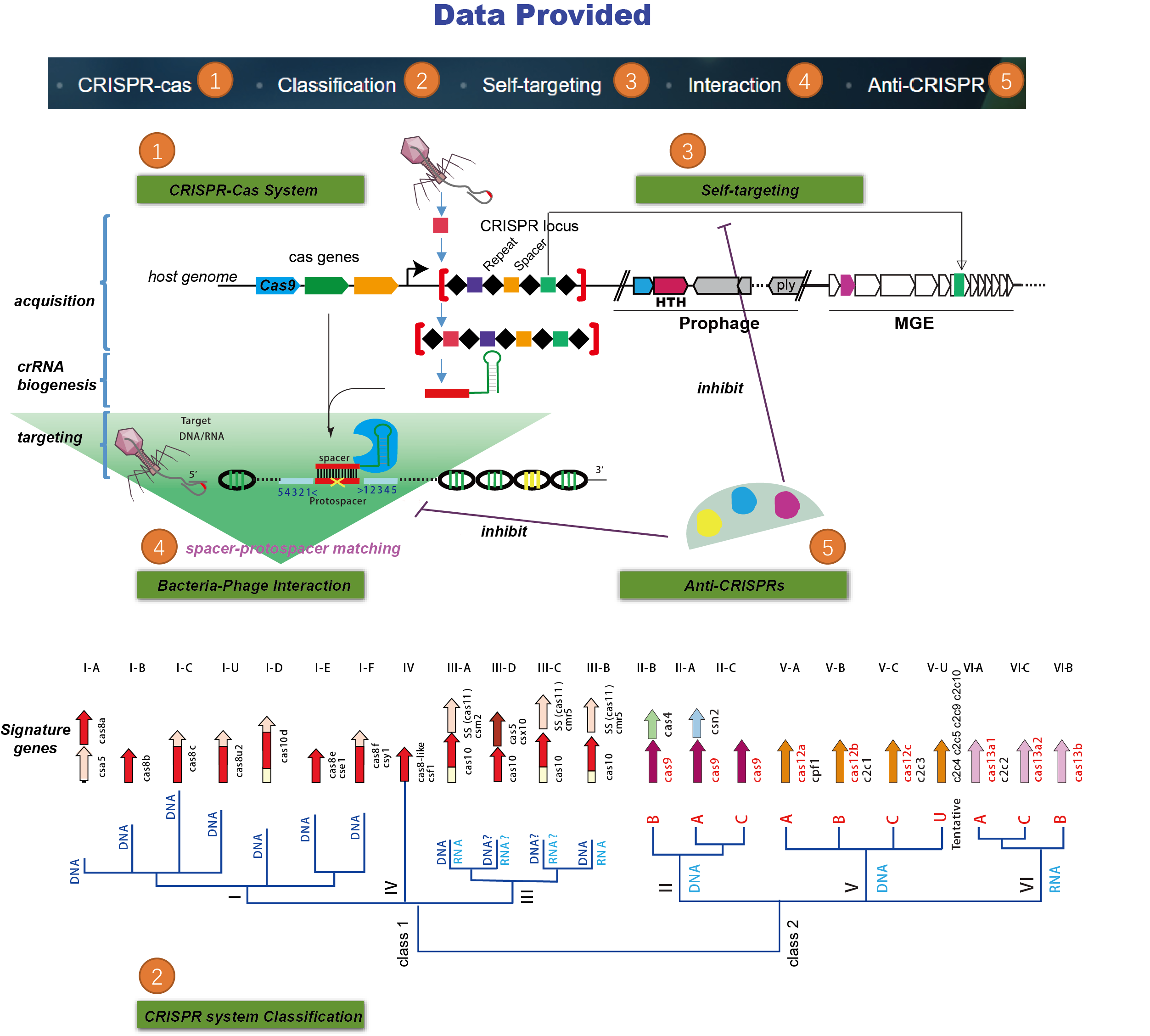
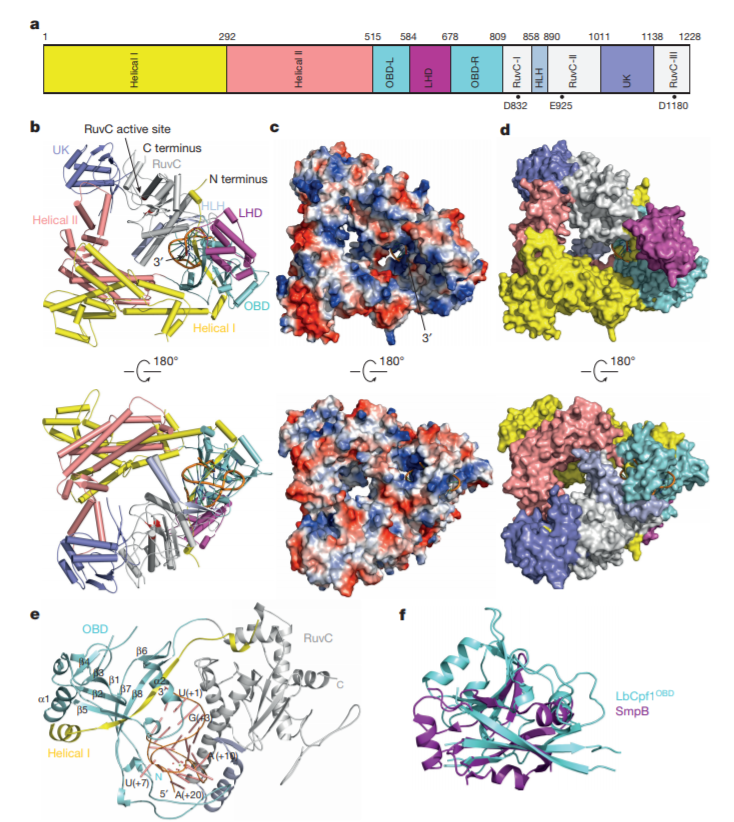
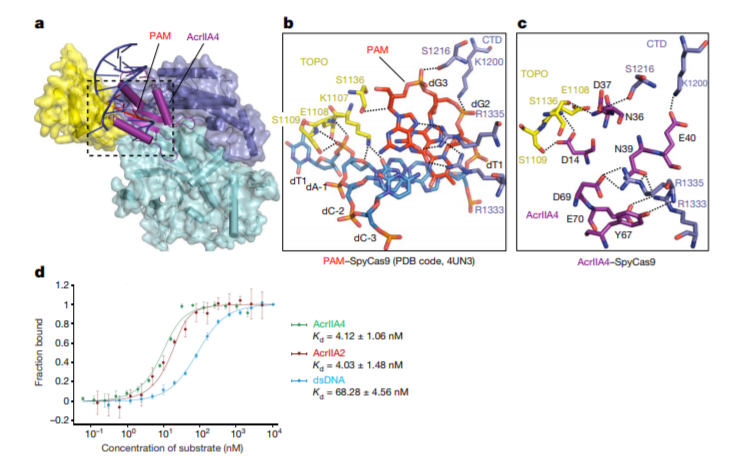
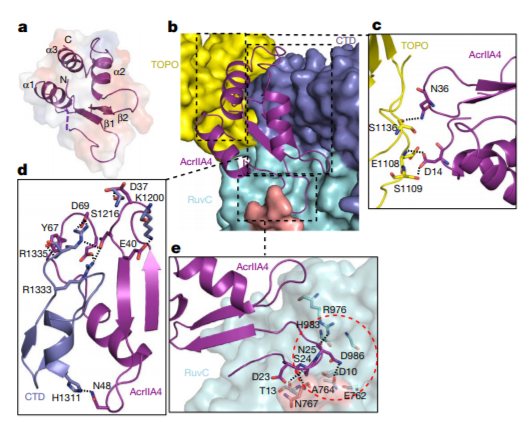
The CRISPR-Cas systems not only play key roles in prokaryotic acquired immunity, but also can be adapted to powerful genome editing tools. Here, we developed CRISPRminer, a knowledge database and web-server to guide CRISPR-Cas associated studies, including CRISPR locus and Cas protein prediction, CRISPR-Cas systems classification, self-targeting detection, microbe-phage infection networks and anti-CRISPRs annotation.