Introduction of CRISPR system
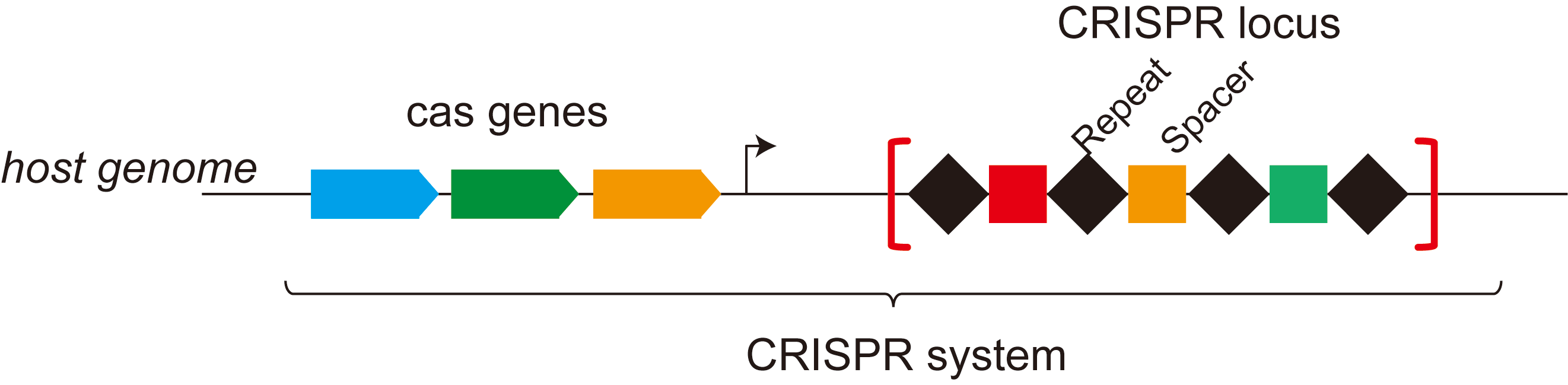
CRISPR: Clustered regularly interspaced short palindromic repeats
Cas: CRISPR associated proteins
CRISPR-Cas system: CRISPR and associated proteins (Cas) comprise the CRISPR-Cas system
CRISPR locus: locus that contains alternate repeated elements (CRISPR repeats) and variable sequences (CRISPR spacers)
The Cas proteins can be divided into four distinct functional modules: adaptation (spacer acquisition), expression (crRNA processing and target binding), interference (target cleavage), and ancillary (regulatory and other CRISPR-associated functions).
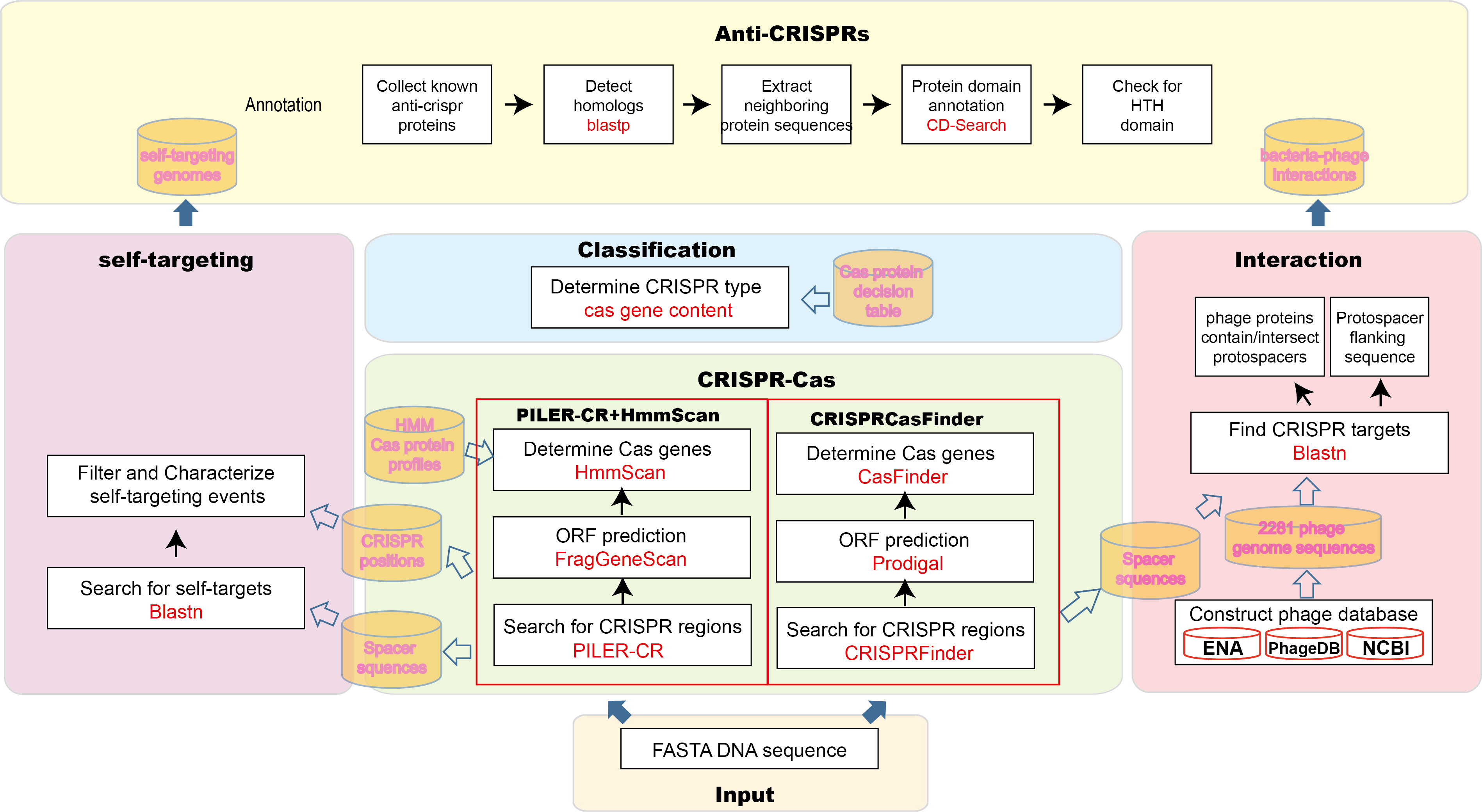
CRISPR system detection work-flow

Browser
Users can browse all the bacterial and archaeal organisms with CRISPR systems (containing both CRISPRs and Cas proteins) according to the bacteria evolutionary lineage.
Compare
All the bacterial organisms with CRISPR systems are organized at the species or genus level. When clicking on a specific species or genus name, all CRISPR systems belonging to the organisms belonging to the selected species or genus are displayed. Thus, users can compare the CRISPR systems to see the common or distinct features among the organisms belonging to the same species or genus.
Visualization of predicted CRISPR-Cas systems
Our system provide both global and local views of predicted CRISPR-Cas system(s). The global view provides predicted bacterial genomic islands (GIs, regions of probable horizontal origin) using IslandViewer and predicted prophage regions using prophinder and phaster for users to compare with the loci of the CRISPR systems.
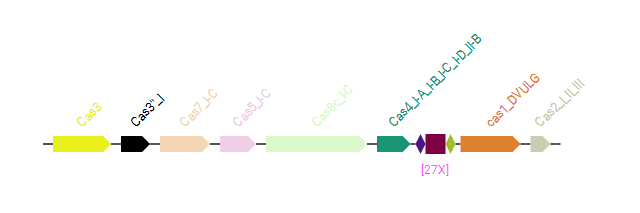
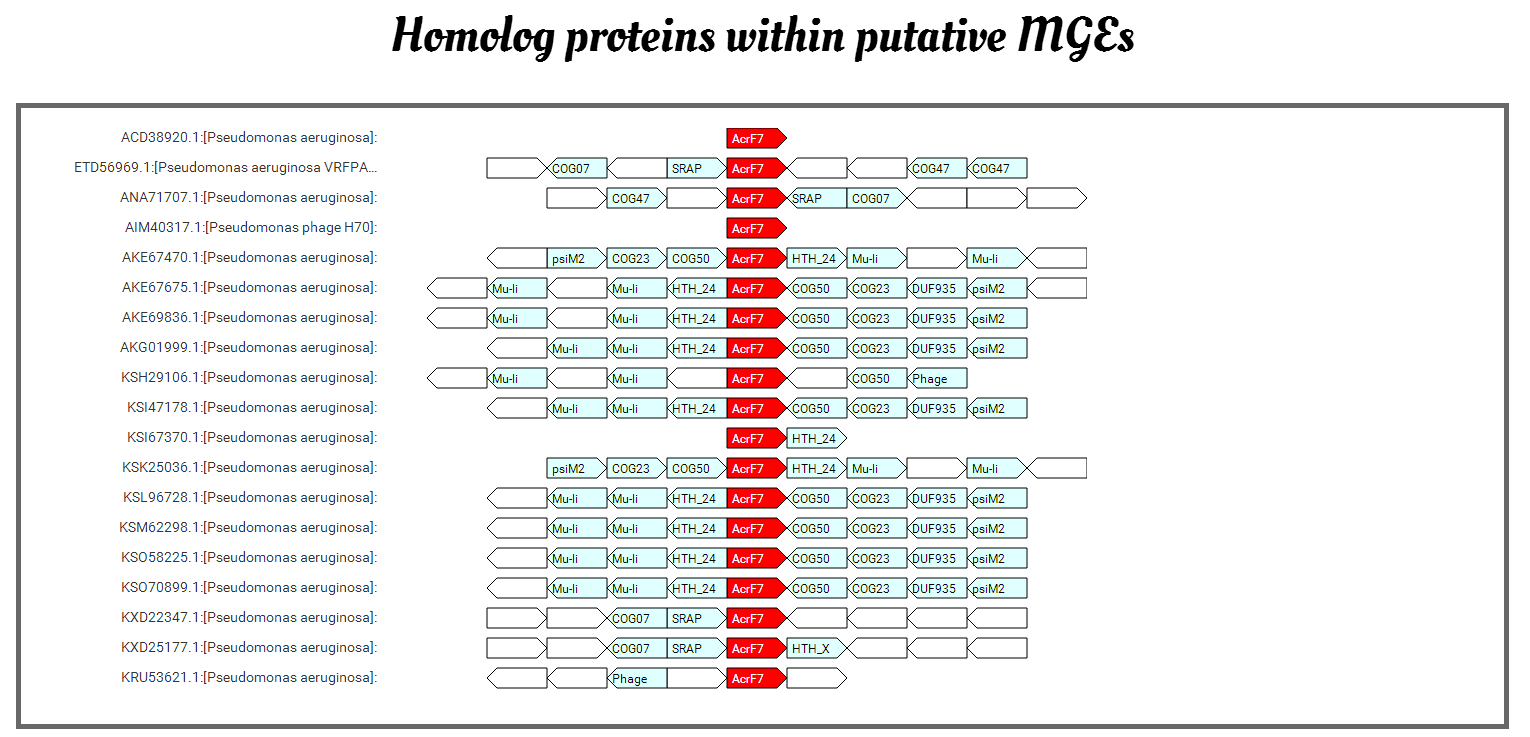
The local view provides the visualization of the CRISPR array (shown as x10 for example, 10 means the copy number of the repeats) plus the Cas proteins (cas proteins in different functional modules are drawn with different colors).
The detailed information for the Cas genes (locations, HMM profiles and type/subtype), the CRISPR array (location, repeat and spacer) are provided at the bottom of the page.