Welcome to CRISPRimmunity!
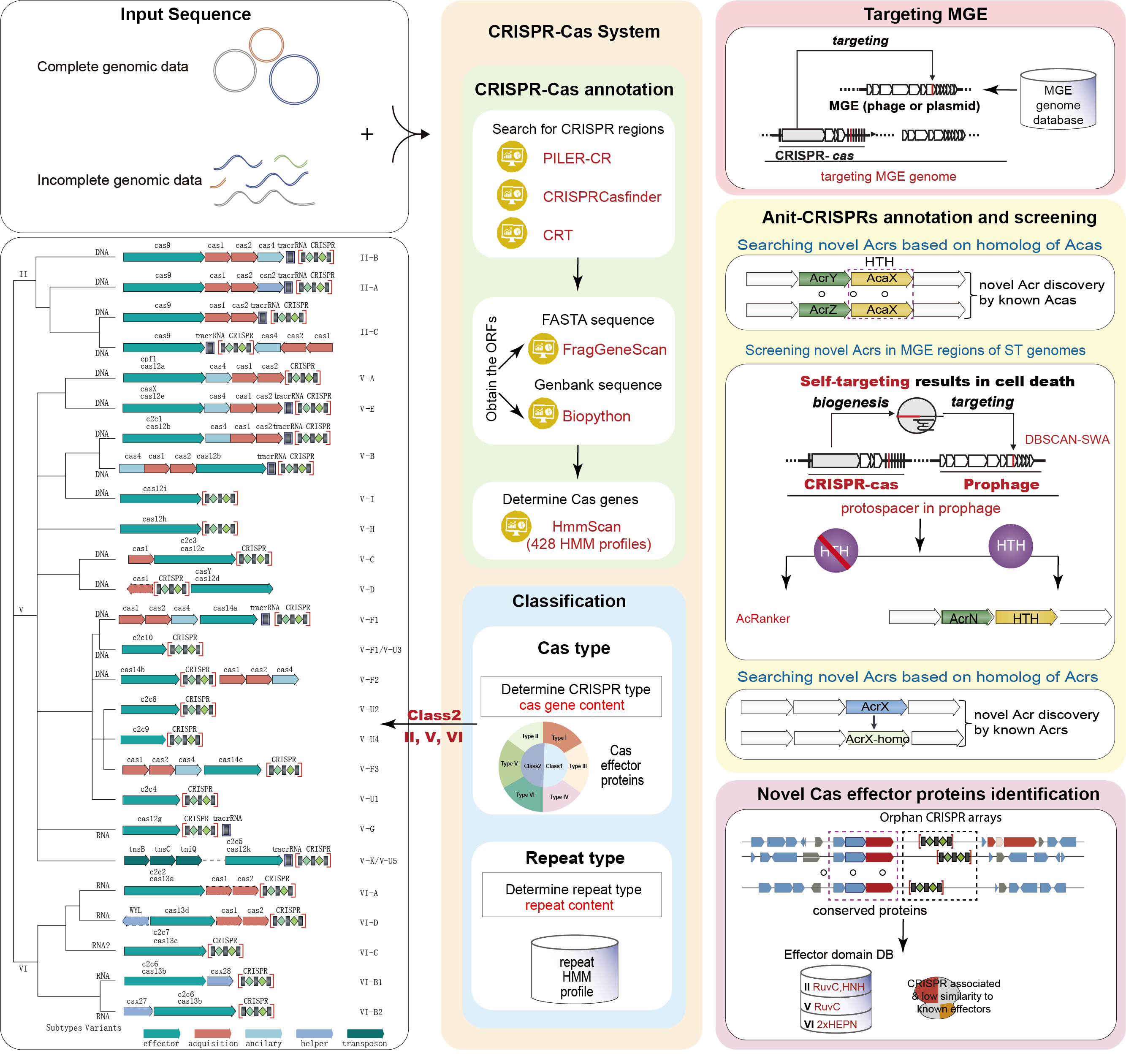
CRISPRimmunity(CRISPR-associated Important Molecular events and Modulators Used in geNome edITing identifY), a CRISPR-oriented web-tool to dissect the key molecular events during the co-evolution of CRISPR and anti-CRISPR mechanisms. CRISPRimmunity incorporated up-to-date knowledge about CRISPR-Cas system detection and annotation, Anti-CRSISPR (Acr) protein and Acr-associated (Aca) gene annotation, and multiple detection options including self-targeting spacer search, repeat type identification, bacteria-phage interaction detection, and prophage detection. CRISPRimmunity achieves enhanced capability in analysis and prediction by applying ‘self-targeting’, ‘guilt by association’ and machine learning approaches for anti-CRISPR protein prediction as well as comparative genomics screening approach for comprehensive identification of novel class 2 CRISPR-Cas loci. The whole prediction tasks are processed remotely by the server using custom python scripts and a series of public tools including CRISPRCasfinder, PILER-CR, CRT, HmmScan, STSS (Self-Targeting Spacer Searcher), DBSCAN-SWA, AcRanker, NCBI Blast+ suite (blastn, blastp, rpsblast) and Diamond. CRISPRimmunity accepts microbial genome data in (multi) FASTA or multi-GenBank file as input, and provides a well-designed graphical user interface and detailed tutorials for input preparation, data processing, and results interpretation.