Welcome1 to PHISDetectorPhage-host interactions are appealing systems to study bacterial adaptive evolution and are increasingly recognized as playing an important role in human health and diseases, which may contribute to novel therapeutic agents, such as phage therapy to combat multi-drug resistant infections. PHISDetector, a web tool to detect and systematically study diverse in silico phage-host interaction signals, including analyses for Sequence composition, CRISPR, Prophage, Genetic homology and protein-protein interactions. PHISDetector uses two criteria to predict phage-host interactions and integrates all Phage-Host Interaction Signals(PHIS) analysis into a PHIE module with machine learning models. PHISDetector accepts various genomics as input and provides well-designed and interactive visualization graphics and detailed data tables to download for each PHIS signal. PHISDetector also provides analysis for oligonucleotide profile/sequence composition,CRISPR-targeting, Prophages, phage genome similarity, special gene check, protein-protein interactions and co-occurrence/co-abundance patterns in Tools. Detect diverse in silico phage-host interaction signals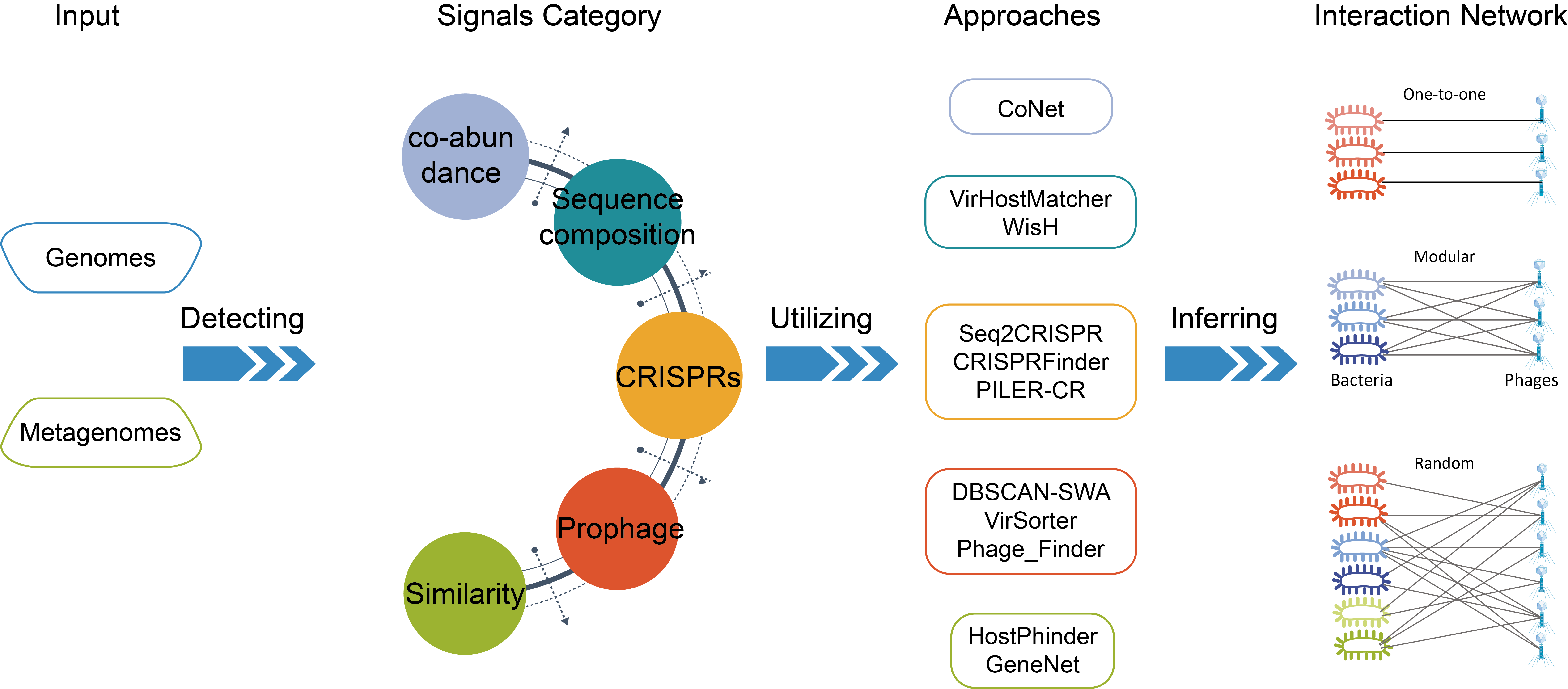
Assess for problematic genes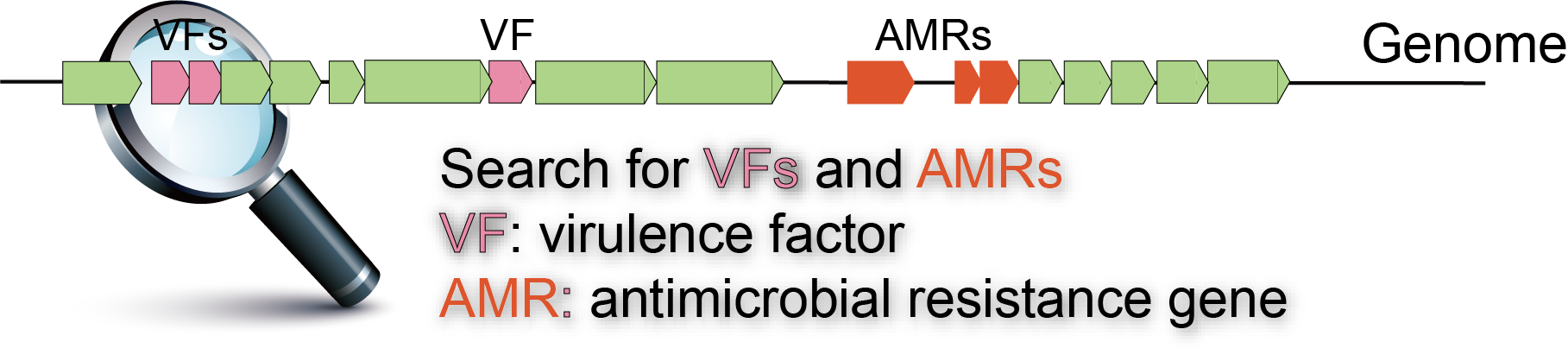
Protein Interaction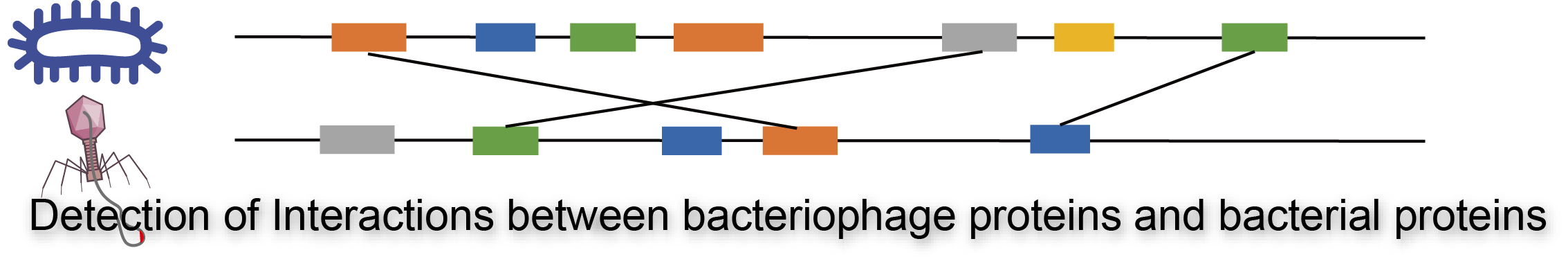
|
Resource LinksCRISPRMiner http://www.microbiome-bigdata.com/CRISPRminer CRISPRCasFinder https://crisprcas.i2bc.paris-saclay.fr/Home/ PILER http://www.drive5.com/pilercr/ WiSH https://github.com/soedinglab/wish VirHostMatcher https://github.com/jessieren/VirHostMatcher HostPhinder https://github.com/julvi/HostPhinder GeneNet https://github.com/coevoeco/GeneNet PHASTER VirSorter https://github.com/simroux/VirSorter Phage_Finder http://phage-finder.sourceforge.net/ ShortBRED https://bitbucket.org/biobakery/shortbred/src RGI https://github.com/arpcard/rgi DIAMOND https://github.com/bbuchfink/diamond BLAST ftp://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST/ FragGeneScan |